Beyond “Junk DNA”: Non-coding RNAs as a Regulatory Layer
For decades, much of the human genome that does not encode proteins was relegated to the category of “junk DNA.” That view has been fundamentally revised. Advances in high-throughput sequencing revealed pervasive transcription across the genome, uncovering a vast landscape of non-coding RNAs (ncRNAs) with regulatory potential.
Importantly, this revision does not imply that all non-coding transcription is functional. While many ncRNAs exhibit tightly regulated, cell-type-specific or stress-responsive expression patterns, others likely represent transcriptional noise or byproducts of RNA processing. Distinguishing functional ncRNAs from incidental transcripts remains critical—particularly when assigning mechanistic roles or proposing diagnostic and therapeutic applications.
Length-Based Classification of ncRNAs: Useful, but Imperfect
Operationally, ncRNAs are commonly classified by length into small ncRNAs (<200 nucleotides) and long ncRNAs (>200 nucleotides). This framework remains widely used and practical, especially in experimental design and bioinformatic pipelines.
However, circular RNAs (circRNAs) challenge this dichotomy. Although often several hundred to several thousand nucleotides long, their defining feature is not size but topology. Circularization confers unique biochemical properties—most notably resistance to exonucleases—that distinguish circRNAs functionally from linear long ncRNAs.
Exosomes as RNA Carriers in the Circulation
Exosomes—small extracellular vesicles of endosomal origin, typically 30–150 nm in diameter—have emerged as important vehicles for RNA transport. Encapsulation within a lipid bilayer protects RNA cargo from degradation by extracellular RNases, enabling remarkable stability in biofluids.
That said, exosomes are not the sole carriers of extracellular RNA. A substantial fraction of circulating RNA is associated with ribonucleoprotein complexes, such as AGO2-bound miRNAs or HDL-associated RNAs. Moreover, while exosome-mediated RNA transfer can be biologically meaningful, the functional impact in recipient cells is highly context-dependent and often quantitatively modest.
How Broad Is the ncRNA Repertoire in Exosomes?
Most major classes of ncRNAs have been detected in exosomes, but their abundance, enrichment, and functional relevance vary widely. Cell type, physiological or pathological state, and technical factors—particularly isolation and sequencing methods—strongly influence observed RNA profiles.
The presence of certain RNA species, such as snRNAs or snoRNAs, may reflect selective export in some contexts, but in others may indicate nuclear RNA turnover, cellular stress, or transformation-associated dysregulation. As such, detection alone should not be equated with function.
Key Classes of Exosomal ncRNAs
MicroRNAs (miRNAs)
miRNAs remain the most consistently enriched and extensively studied exosomal RNA species. Their sorting into exosomes is regulated by RNA-binding proteins such as hnRNPA2B1, YBX1, and SYNCRIP, often recognizing specific sequence motifs known as “EXOmotifs.”
While exosomal miRNAs can modulate gene expression in recipient cells, claims of rapid or robust effects should be tempered. Biological impact depends on multiple factors, including miRNA copy number per vesicle, uptake efficiency, and effective loading into the RNA-induced silencing complex (RISC).
Long Non-coding RNAs (lncRNAs)
Tumor-derived exosomes are frequently enriched for specific lncRNAs, including well-studied examples such as HOTAIR, MALAT1, and H19. Experimental evidence supports roles for some exosomal lncRNAs in fibroblast reprogramming, angiogenesis, and immune modulation.
Nevertheless, the strength of causal evidence varies across models. While certain mechanisms are well supported, many reported associations remain correlative, underscoring the need for functional validation.
Circular RNAs (circRNAs)
circRNAs are particularly stable in extracellular environments, contributing to their enrichment in exosomes and their appeal as biomarkers. In addition to nuclease resistance, active export of circRNAs may serve as a cellular clearance mechanism.
Although circRNAs are often described as miRNA sponges, this function is highly context-specific and frequently overstated. In many cases, circRNA abundance is insufficient to exert meaningful stoichiometric effects. Alternative functions—including protein binding, transcriptional regulation, and even translation into micropeptides—are increasingly recognized.
tRNA-Derived Fragments (tRFs)
tRNA fragments and stress-induced tiRNAs are among the most abundant small RNAs found in exosomes across multiple biofluids. Far from being random degradation products, these molecules have been implicated in translational repression, stress signaling, metabolic regulation, and immune modulation.
Their enrichment in exosomes likely reflects a combination of regulated export and increased cleavage under cellular stress.
Additional ncRNA Species
Beyond the commonly highlighted classes, exosomes also contain:
Y RNAs and Y RNA fragments, particularly abundant in plasma extracellular vesicles
Vault RNAs
Repeat-derived RNAs
These species are gaining attention as contributors to extracellular vesicle RNA signatures and may carry diagnostic or functional significance.
Exosomal ncRNAs and Liquid Biopsy Applications
Exosomal ncRNAs are actively explored as biomarkers for cancer, neurodegenerative diseases, and other conditions. Their stability and disease-associated expression patterns make them attractive candidates for liquid biopsy approaches.
However, several challenges remain: lack of standardization in vesicle isolation, contamination by non-vesicular RNA, and significant inter-individual variability. While clinical translation is advancing, relatively few assays have yet achieved validation for routine clinical use.
A Balanced Perspective
Exosomes harbor a diverse but non-random repertoire of non-coding RNAs—including miRNAs, circRNAs, lncRNAs, tRNA-derived fragments, and other small RNAs—whose composition reflects both selective sorting mechanisms and the physiological or pathological state of the originating cell. This complexity makes exosomal ncRNAs compelling subjects for intercellular communication research and biomarker discovery, while also demanding methodological rigor and biological restraint in interpretation.
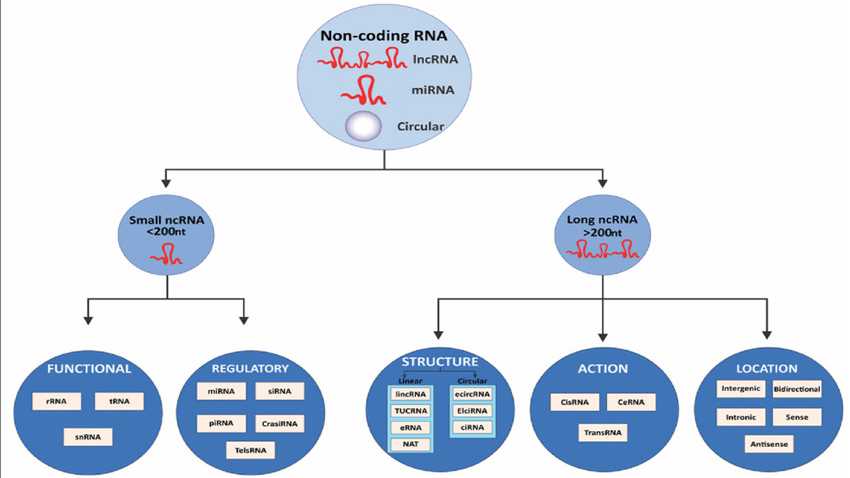
Classification of noncoding RNAs (ncRNAs)
https://www.researchgate.net/figure/Classification-of-noncoding-RNAs-ncRNAs-Noncoding-RNAs-are-classified-into-small_fig1_339894753